Part:BBa_K2127002
CP47 His-tagged with Lac Promoter
The psbB gene codes for the CP-47 subunit of the Cyanobacterial Photosystem II in Synechocystis sp. PCC 6803. Using site-directed mutagenesis Dr Frankel’s team developed a Synechocystis sp. PCC 6803 mutant containing a histidine tag at the C-Terminus of CP47 subunit. Using the His-Tag, the Photosystem II complex can be isolated and analyzed the activity using an electron evolution analyzer.
BBa_K2127002 is a composite part which contains the coding region of the psbB with an added his-tag and a double stop codon. The composite site also contains a lac promotor, RBS,and terminator which allow for direct expression of this insert. Assembly was done via sirect synthesis from IDT technologies as a gBlock. As a result no scar sites exist within the composite part. The BioBrick is able to be used directly with E. coli for protein expression and can also be transitioned into cyanobacterium with some genetic work for expression of His-tagged Photosystem II complexes.
Usage and Biology
The his-tagged CP47 subunit is approximately 48kDa and is a transmembrane protein. As part of the Photosystem II complex, the subunit binds chlorophyll and acts as part of the light capturing array. The subunit ligand sites allow for electron movement across the peptide to facilitate movement within the electron transfer chain from both the light capturing array and from the photolysis of water. The subunit also composes part of the manganese calcium complex for oxygen evolution. It is extremely integral to the function of the PSII complex.
Also in conjunction to its functions, CP47 also forms part of the quinone channel which allows direct transfer of electrons from the D1 monomer to quinones of the electron transport chain.
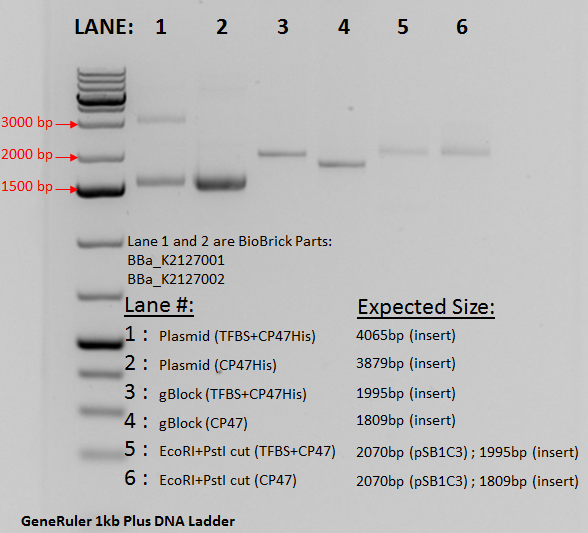
Verification
A plasmid containing the composite part was successfully made and transformed into E. coli DH5aF'Iq. The plasmid was extracted using miniprep and double digested using EcoRI and Psti. The gel below confirms the transformation. Lane 2 contains the undigested plasmid running at approximately 1500 bp due to a supercoiled nature. Lane 4 contains the gBlock from IDT and acts as a size control. The band is seen aa expected at 1809 bp. In lane 6, the digested plasmid is seen. Due to both the pSB1C3 vector and the psbB insert being very close to each other at 1809bp and 2070 bp interference did not allow for a clear separation of the bands. However compared to Lane 2 it can bee seen that digestion did occur since the run time changes significantly between the digested linear sample and the undigested supercoiled plasmid.
Functionality
The composit part BBa_K2127002 was also tested for functionality. The lac promoter allows for inducement using IPTG and the His tag allows for purification of the protein. The figure shows that expected band of our CP47His protein at 48kDa. Since lac is a leaky promoter the darkening of the band in the cell lysate is expected. The flow through and the washes also contain our band wih increasing clarity. The inability for the his-tag to bind properly to the column was due to the use of a protein inhibitor cocktail that contained EDTA. The EDTA caused chelation with the nickel ligands and resulted in our protein washing out with subsequent washes. The gel does confirm the the BBa_K2127002 is being successfully expressed according to our design.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 473
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 597
Illegal AgeI site found at 1194
Illegal AgeI site found at 1398 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 918
Illegal SapI site found at 1595
//cds/enzyme
//cds/membrane
//chassis/prokaryote/cyanobacterium
//plasmid/expression
| chassis | Synechocystis spp. PCC 6803 |
| family | Photosystem II Reaction Complex |
| function | Light Harvesting, Oxygen Reaction Complex Stabilization |
| insert | Expressable |
| ligands | Histine, Fe(II), Chlorine, Mangenase, Calcium (II), Chlorophyll, Beta-Carotene |
| protein | 47 kDa subunit |
| tag | Nickel Affinity Histine |